Protein design workshop at PSI in Switzerland
Published:
Ajsaja was invited to lead a two-day protein design workshop at the Paul Scherrer Institute (PSI) in Switzerland. He was joined by Federico and Eva as teaching assistants.
Published:
Ajsaja was invited to lead a two-day protein design workshop at the Paul Scherrer Institute (PSI) in Switzerland. He was joined by Federico and Eva as teaching assistants.
Published:
Our lab participated in the European Conference on Protein Design, which brought together leading scientists in the field for a dynamic exchange of ideas and discoveries. Liza and Eva showcased their research through poster presentations, while Alina was selected to give a lecture on her work involving de novo designed symmetrical assemblies.
Published:
Ajasja and Eva visited the Nobel Prize ceremony in Stockholm, Sweden 🇸🇪.
More on rosettacommons blog
Published:
Federico attended the cryo-EM sample preparation workshop at the ESRF in Grenoble.
Published:
Our laboratory was granted 25000 hours on the VEGA cluster. We will be using this processing power to work on designs for fiber anchors, rigid walkers and more!
Published:
We are excited to announce that our lab will be hosting RosettaCon Europe 2025 in Ljubljana from October 20–22, 2025. This event will bring together researchers from across the Rosetta community to discuss the latest advances in computational biology and protein design. Stay tuned – registration will open soon! Link to website: https://europeanrosettacon.org/
Published:
Eva was awarded a travel grant to attend Summer RosettaCon in August. Congratulations Eva!
Published:
Ajasja, Liza and Federico attended the ArtMotor consortium meeting as invited guests, where Ajasja presented our group’s advancements in de novo designed motors
Published:
We would like to welcome our first Rosetta Commons REU student Kiyan Abel!
Published:
We are happy to host Nej Bizjak for the summer. Among other things he will work on improving Prosculpt.
Published:
Even in the age of Slack and Zoom, a happy hour is the best information transfer and ideas catalyst.
Published:
A proud lab moment: our very first master’s student defense is complete!
Published:
We released an updated version of prosculpt:
Published:
Vacations are over and proteins are waiting to be designed!
Published:
Lab milestone reached – we now have lab T-shirts! :)
Published:
We’re very happy to announce our new paper is in preprint! You can read it at this link
Published:
In the PROPEL project we will create a completely de novo designed powered protein walker. More info here.
Published in J. Am. Chem. Soc., 2017
The coiled-coil dimer is a widespread protein structural motif and, due to its designability, represents an attractive building block for assembling modular nanostructures, which we demonstrated in 2013 in a paper in Nat.Chem.Biol. The specificity of coiled-coil dimer pairing is based on hydrophobic and electrostatic interactions. We showed how design of the local helical propensity of interacting peptides can be used to tune the stabilities of coiled-coil dimers over a wide range. This general principle was demonstrated by a change in thermal stability by more than 30 °C as a result of only two mutations outside the binding interface. Our findings enable a diverse range of applications in synthetic biology and nanomaterials. This study by authors Igor Drobnak, Helena Gradišar, Ajasja Ljubetič, Estera Merljak and Roman Jerala was published in the paper “Modulation of Coiled-Coil Dimer Stability through Surface Residues while Preserving Pairing Specificity” in the Journal of the American Chemical Society
Published in Current opinion in chemical biology, 2017
Advances in design of protein folds and assemblies
Published in Current opinion in chemical biology, 2017
Advances in design of protein folds and assemblies
Published in Nature biotechnology, 2017
Proteins are nature’s nano-robots performing key processes necessary for sustaining life. They are built as a linear chain of amino acids which encodes information about the 3D structure and their function. The number of possible sequences is practically infinite: all possible combinations of a 64 amino acid long protein exceed the number of atoms in the known universe. Design of novel proteins is also hampered by extremely complex folding rules.
Published in Nature biotechnology, 2017
Proteins are nature’s nano-robots performing key processes necessary for sustaining life. They are built as a linear chain of amino acids which encodes information about the 3D structure and their function. The number of possible sequences is practically infinite: all possible combinations of a 64 amino acid long protein exceed the number of atoms in the known universe. Design of novel proteins is also hampered by extremely complex folding rules.
Published in Nat Chem Biol, 2018
Activity of genes is defined by proteins that bind to the DNA in their close proximity. Transcription Activator-Like Effector (TALE) proteins can be designed to bind almost any selected DNA sequence. Researchers at the Department of Synthetic Biology and Immunology at NIC reported several years ago that TALEs can be used to introduce logic functions into mammalian cells. Now researchers from the same group report in an article in Nature Chemical Biology that TALE proteins can displace other TALE proteins from the DNA in a highly polarized manner –the displacement of a TALE can occur only by binding of another TALE protein to the DNA sequence adjacent to its left but not its right side. Based on a chained displacement of several adjacent TALE proteins, the researchers managed to implement all two-input logic circuits in human cells.
Published in Org. Biomol. Chem., 2019
Published in Nat Commun, 2021
Published in Nat Commun, 2021
Published in Nature machine intelligence, 2022
Machine learning with deep neural networks has accelerated research and applications in many areas, from translating texts, playing chess, to designing new proteins that can serve as drugs and vaccines. An example of a successful neural network is DeepMind’s AlphaFold2. Difficulties arise in interpreting neural networks. The answer to the question “Why and how did the network offer us a certain answer?” is difficult or unknown.
Published in Nature machine intelligence, 2022
Machine learning with deep neural networks has accelerated research and applications in many areas, from translating texts, playing chess, to designing new proteins that can serve as drugs and vaccines. An example of a successful neural network is DeepMind’s AlphaFold2. Difficulties arise in interpreting neural networks. The answer to the question “Why and how did the network offer us a certain answer?” is difficult or unknown.
Published in Chem, 2022
Structurally complex and dynamic biomolecular assemblies are key components of lifelike systems and materials. State-of-the-art rules and guiding principles for the design of these peptide-, protein-, and polynucleotide-based nanostructures were discussed by a group of international leading scientists gathered at the virtual EMBO workshop.
Download here
Published in Science Translational Medicine, 2022
Highlights:
Published in Nature Communications, 2023
Highlights:
Published in Nature Communications, 2023
Highlights:
Published in Cell Discovery, 2024
Schematic illustration of the structure engineered molecular scissors Cas9 regulated through binding of the peptide to the insert (red).
Published in Cell Discovery, 2024
Schematic illustration of the structure engineered molecular scissors Cas9 regulated through binding of the peptide to the insert (red).
Published in J. Phys. Chem. B, 2024
Published in Nature chemical biology, 2024
Highlights:
Published in Nature chemical biology, 2024
Highlights:
Published in ACS Nano, 2024
Modular protein engineering is a powerful approach for fabricating high-molecular-weight assemblies and biomaterials with nanoscale precision. Herein, we address the challenge of designing an extended nanoscale filamentous architecture inspired by the central rigid rod domain of human dystrophin, which protects sarcolemma during muscle contraction and consists of spectrin repeats composed of three-helical bundles.
Published in Angewandte Chemie International Edition, 2025
Until now, scientists have used complementary dimerization modules of coiled coils to design modular protein structures, which they have called protein origami. Researchers from the Department of Synthetic Biology and Immunology have expanded the toolkit for modular nanostructure building blocks by using tetramerization modules instead. The dimeric building blocks were based on pairs of coiled coils, while the tetrameric building blocks consist of four helices that can orient parallel or antiparallel and bundle together.
Published in Current Opinion in Biotechnology, 2025
Recent advances in protein engineering have revolutionized the design of bionanomolecular assemblies for functional therapeutic and biotechnological applications. This review highlights the progress in creating complex protein architectures, encompassing both finite and extended assemblies. AI tools, including AlphaFold, RFDiffusion, and ProteinMPNN, have significantly enhanced the scalability and success of de novo designs. Finite assemblies, like nanocages and coiled-coil-based structures, enable precise molecular encapsulation or functional protein domain presentation. Extended assemblies, including filaments and 2D/3D lattices, offer unparalleled structural versatility for applications such as vaccine development, responsive biomaterials, and engineered cellular scaffolds. The convergence of artificial intelligence–driven design and experimental validation promises strong acceleration of the development of tailored protein assemblies, offering new opportunities in synthetic biology, materials science, biotechnology, and biomedicine.
Published in Current Opinion in Biotechnology, 2025
Recent advances in protein engineering have revolutionized the design of bionanomolecular assemblies for functional therapeutic and biotechnological applications. This review highlights the progress in creating complex protein architectures, encompassing both finite and extended assemblies. AI tools, including AlphaFold, RFDiffusion, and ProteinMPNN, have significantly enhanced the scalability and success of de novo designs. Finite assemblies, like nanocages and coiled-coil-based structures, enable precise molecular encapsulation or functional protein domain presentation. Extended assemblies, including filaments and 2D/3D lattices, offer unparalleled structural versatility for applications such as vaccine development, responsive biomaterials, and engineered cellular scaffolds. The convergence of artificial intelligence–driven design and experimental validation promises strong acceleration of the development of tailored protein assemblies, offering new opportunities in synthetic biology, materials science, biotechnology, and biomedicine.
Published in BioArxiv, 2025
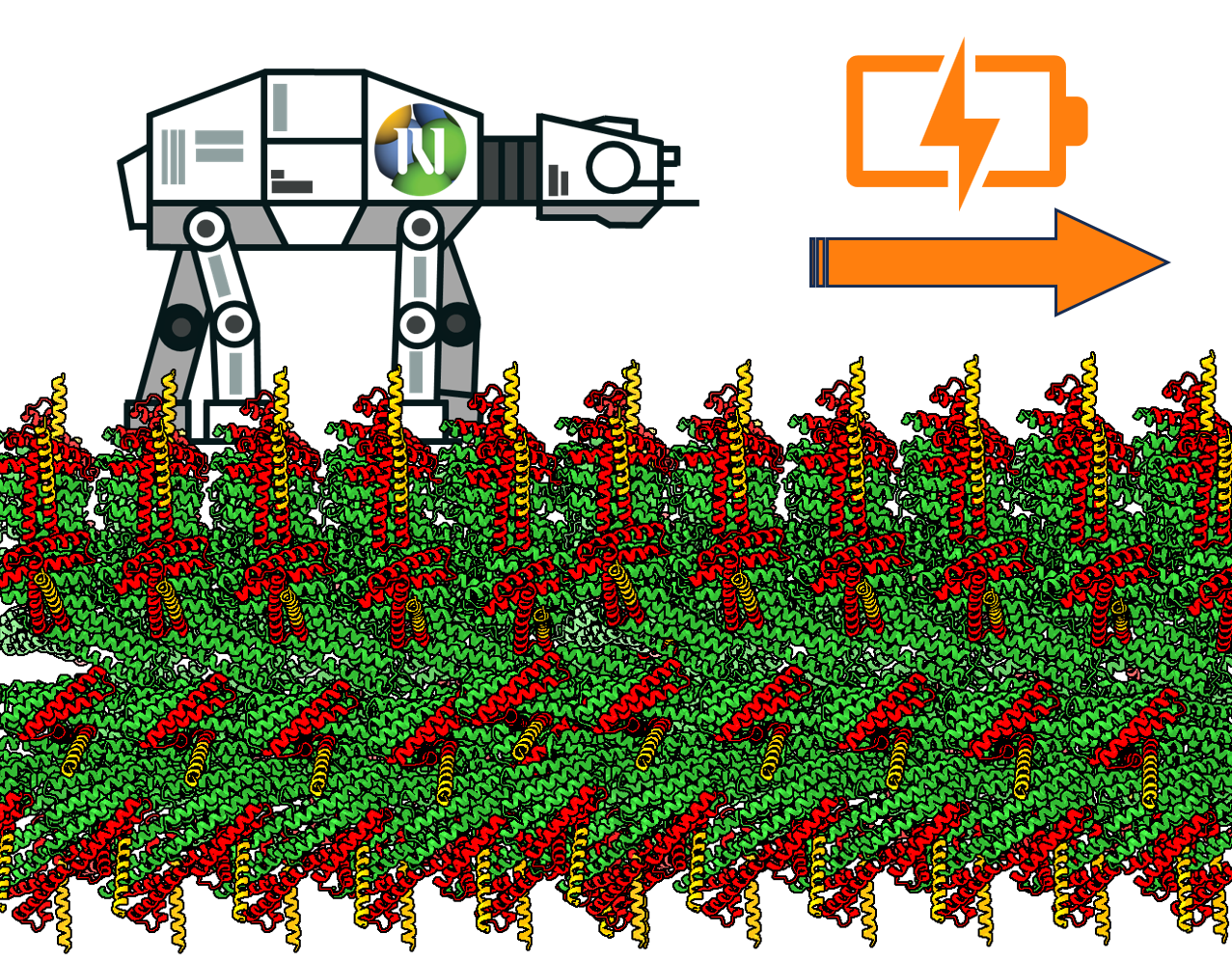
Molecular machines hold great potential. Design of static monomeric and oligomeric protein structures has advanced tremendously, but few dynamic protein systems have been designed. Here we present the design and characterization of a random protein walker that diffuses along a designed protein track. For the track, we designed micro-meter long fibres and developed a method to rigidly decorate them with arbitrary proteins. The walkers consist of homo-oligomers that reversibly bind the track using heterodimeric feet; we tested multiple heterodimer interfaces for reversibility and designed six walkers with different numbers of feet. Cryo-EM experiments confirmed the structure of the track and walkers. We performed detailed single molecule tracking and kinetics characterisation and found that the walkers diffuse along the track, and that those with more feet diffuse faster. The system represents a tuneable starting point for future powered protein molecular robots.
Published:
Published:
Proteins are nature’s nano-robots, catalysing reactions, recognising molecules, and transporting cargo. Biomimetic nanomachines have great potential in many fields including precision medicine, therapeutic agents, nanomaterials, and sensors, however use is limited by inadequate stability of natural protein components. De novo designed proteins are hyper-stable and can achieve shapes and properties not found in nature.
Published:
Protein design and sculpting using Rosetta and Deep learning methods (RFDiff and Alphafold2)
Download here
Design platform for creating single-chain polyhedral cages made from coiled-coil building modules
Download here
Scripts to align various channels based on reference beads.
Download here
Kiyan worked on rigid walkers with us as part of the Rosetta Commons REU
Alina is designing radially symmetric oligomeric proteins to use as anchors of designed fibers
The vision for my research group is to design and study mechanic protein assemblies capable of producing work and make the first steps towards designer nanorobots.
Federico works in the de novo design of protein fibers and molecular motors
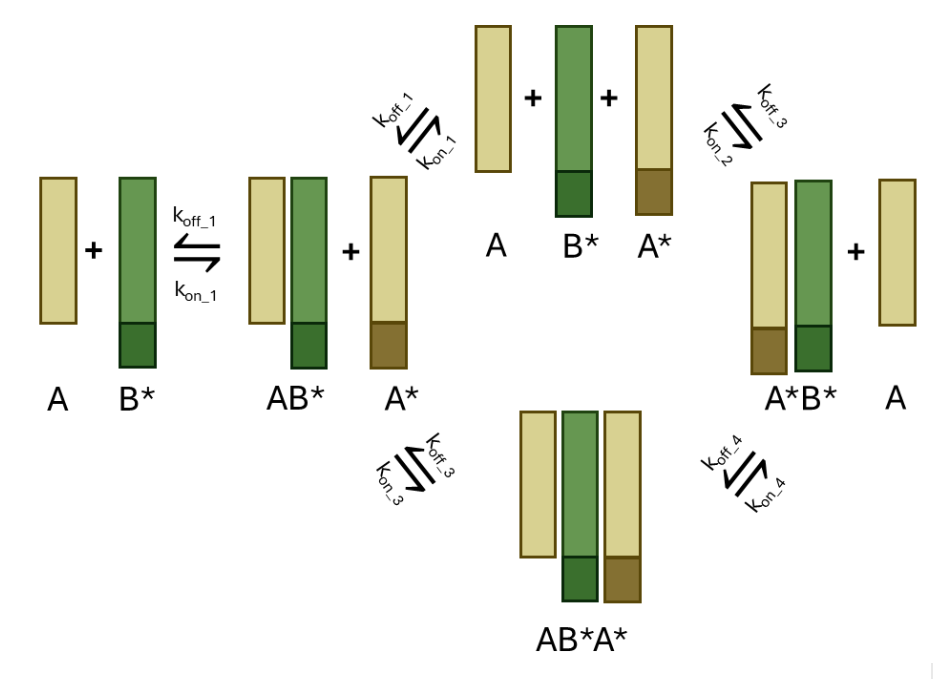
Eva’s work focuses on implementation and characterisation of Toehold Mediated Strand Displacement in proteins
Neža works on the design of symmetric protein anchors
Teo works developing electric microscope cells
The main focus of Liza’s work is characterization of de novo designed molecular walkers
Enej works on the kinetics of protein displacement
Combining the wonders of biochemical world with the usefulness of programming.