Research fields
De novo design of molecular machines
We use cutting edge protein design methods (RFDiffusion, ProteinMPNN, Alphafold2/3) to design scaffold for protein-based molecular machines. Our main focus is building programmable protein walkers, capable of walking on designed tracks. CC WALK lays the foundation for new types of synthetic, controllable systems at the molecular level.
De novo design of coiled coils and four helix bundles
We specialize in designing coiled coils and four-helix bundles from scratch, with precise control over orthogonality, binding affinity, and interaction kinetics. We have discovered new protein design rules for orthogonality (Barshev et al.), created the largest set of orthogonal coiled coils (Boldridge et al.), built intricate protein origami structures (Ljubetič et al.), and shown how coiled coils can regulate enzyme activity (Plaper et al.).
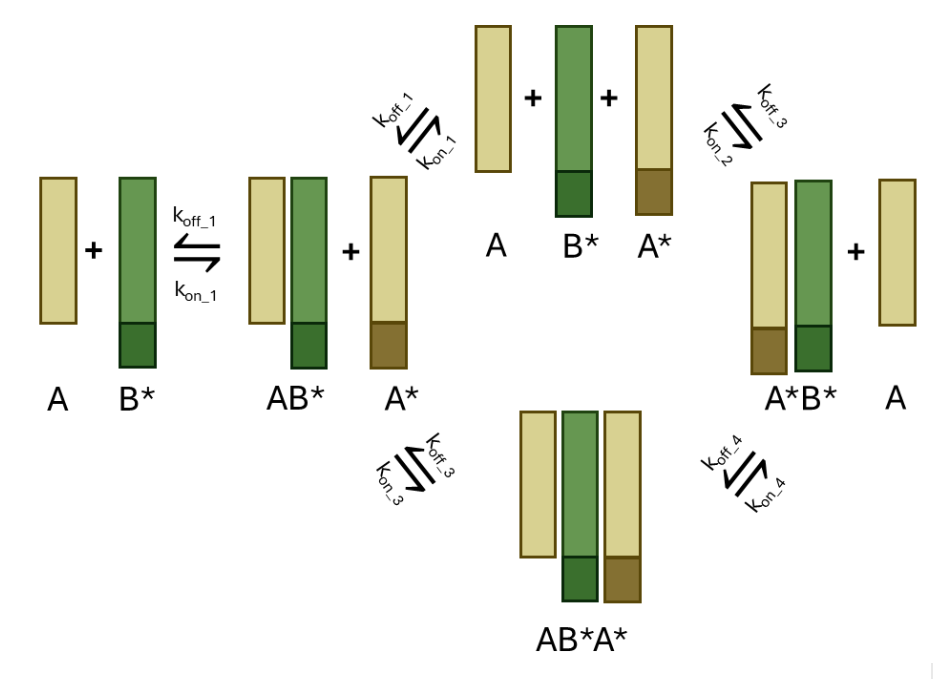
Design of strand displacement proteins
We’re exploring how to adapt the powerful concept of DNA strand displacement to proteins. This approach could offer dynamic control over protein kinetics and open the door to reconfigurable protein-based machines. Learn more in our CC-Trigger project.
Design of mini binders
We engineer custom proteins that bind tightly and specifically to natural targets, such as viral proteins, receptors, or enzymes. We use Prosculpt (RFDiffusion + proteinMPNN) and Bindcraft.